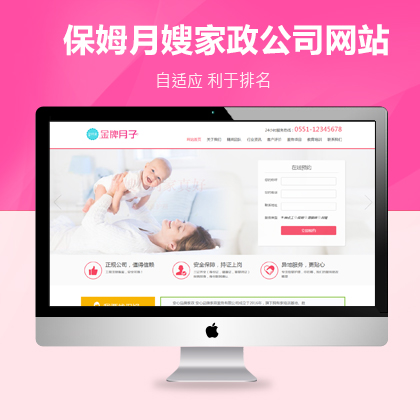
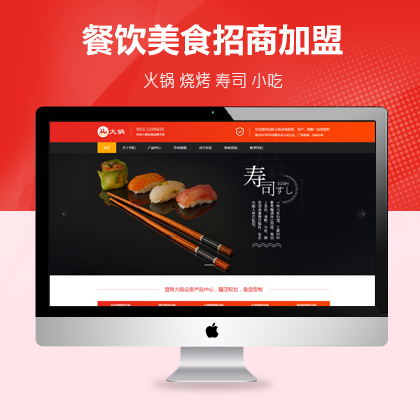
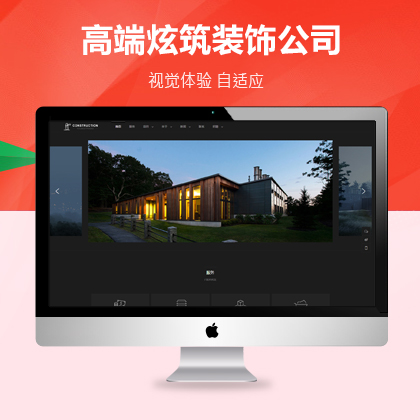
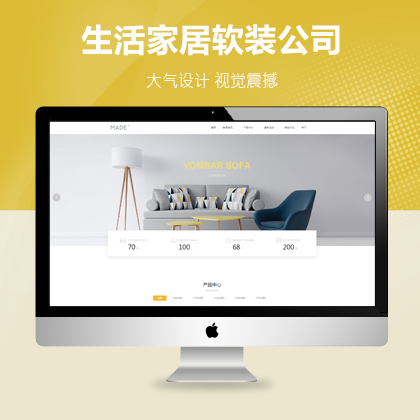
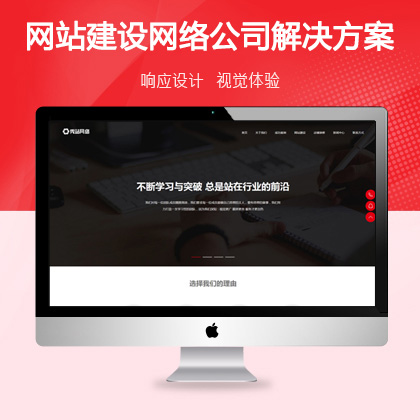
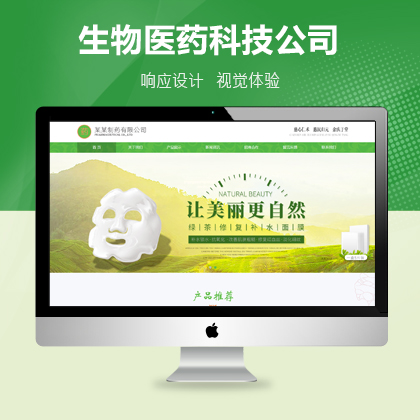
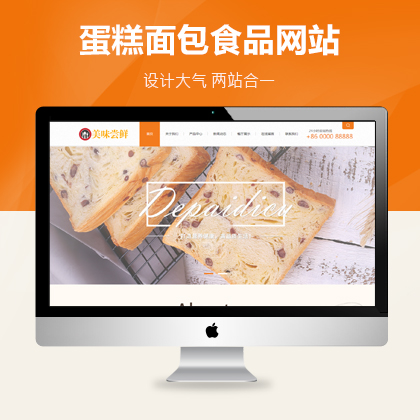
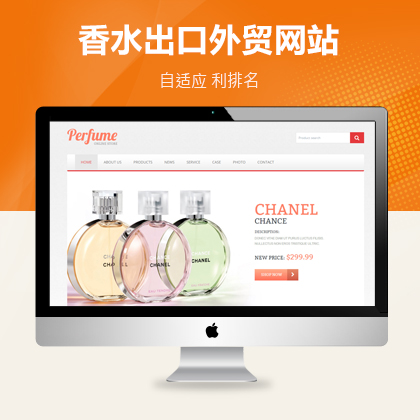
设计师一对一量身设计,定制网站仅1280元起
精选网站建设套餐,打造以量身创意为核心的网站建设,为客户提供一站式网站建设解决方案
-
看肏屄视频播放
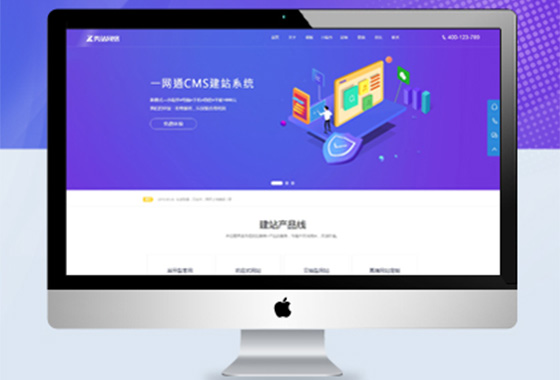
功能型网站顾名思义,就是带有不同属性
用途更广,功能跟强大,满足企业的需求
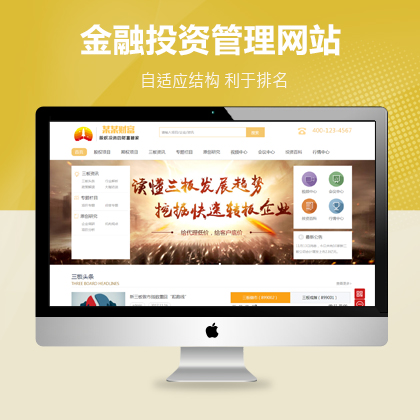
成熟品牌公司,就选功能型网站定制开发¥ 12000.00了解详情
-
干逼污污
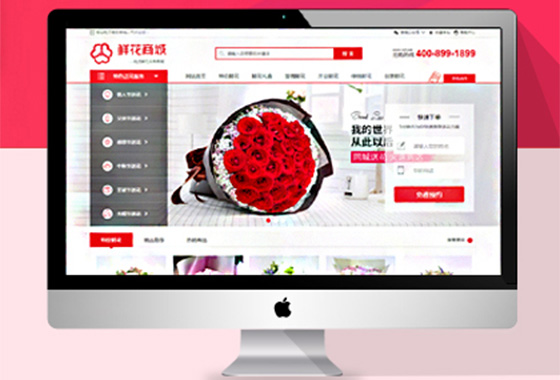
定制开发商城网站个性化网站,高效稳定
轻松帮您管理客户,管理订单,管理商品
助力商家实现销量变现,为店铺带来客户¥ 6800.00了解详情
-
逼逼穴操逼逼逼逼穴
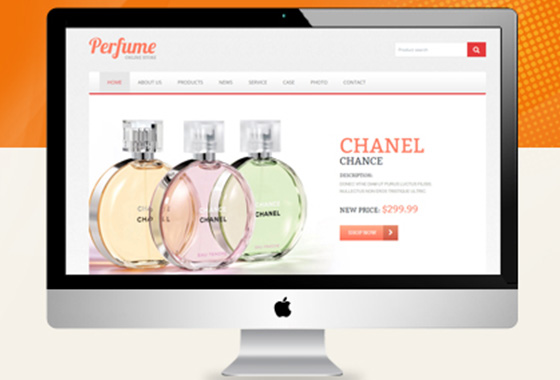
涉及外贸行业的企业,想获得互联网市场
那么必须要有一个能够吸引人的外贸网站
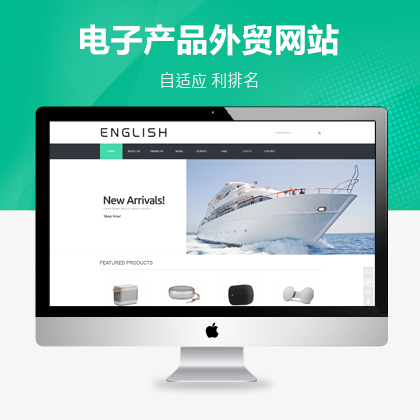
聚为科技提供高端外贸型网站定制化服务¥ 4680.00了解详情
-
毛逼爱流水
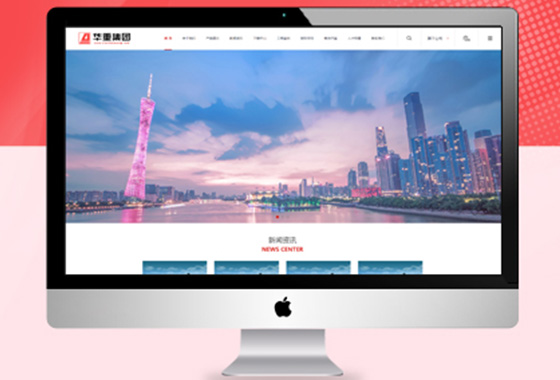
集团公司的业务、服务、产品丰富多样化
定制型网站建设科技满足集团企业的需求
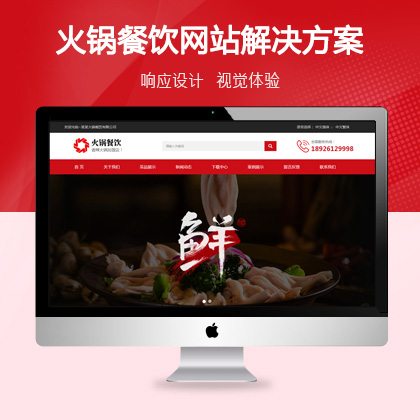
在网站策划和视觉效果设计方面尤为重要¥ 2800.00了解详情
-
美女的小穴
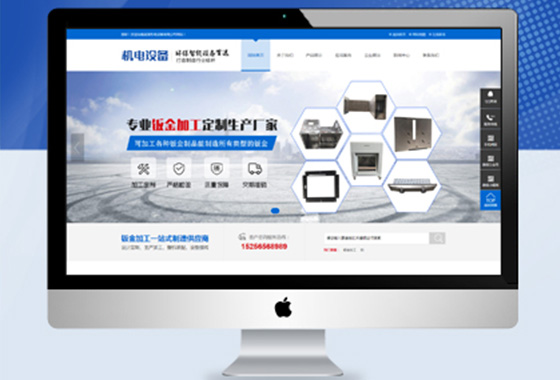
所有导航栏目页面设计,网站更具吸引力
专为网站手写定制程序,百度更容易收录
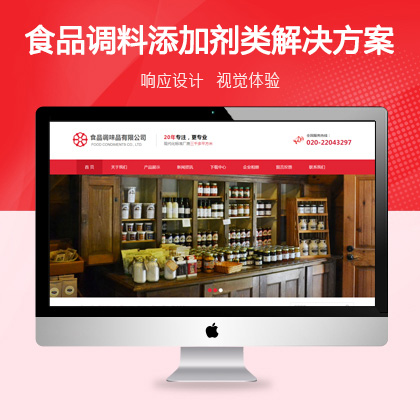
可以设置网站关键词及描述专为排名优化¥ 3680.00了解详情
-
肏逼视频插
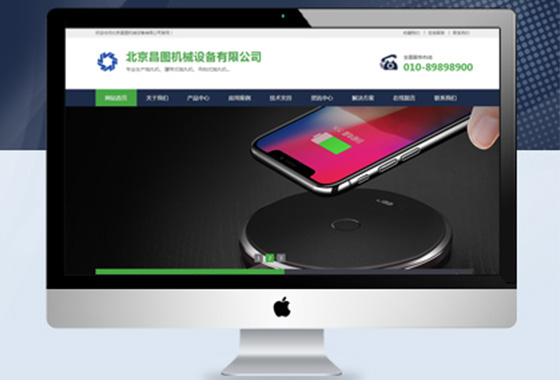
以营销为主导向,让品牌网站更有意义
作为一家企业,想要做大做强网络营销
须拥有一个高品质营销型网站宣传企业¥ 2800.00了解详情
-
极品白丝白虎被强行插入鸡巴
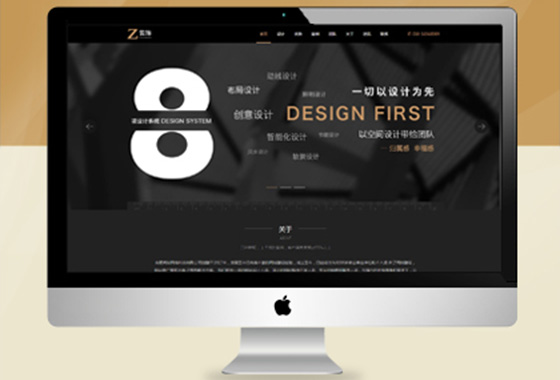
专注网站定制,网站建设,针对各行各业
专业原创网站设计,送网站域名空间备案
适合初期创业个人企业,以展示企业形象¥ 2680.00了解详情
-

成人骚逼av
专用汽车网站定制,拒绝千篇一律的网站
放眼望去,各种专汽网站打开都是一样的
让用户毫无体验感,降低用户信赖与访问¥ 1680.00了解详情
客户案例,坚持原创设计做有价值的网站
专注网站设计,app开发,小程序开发,为企业提供互联网+的基础建设及营销服务
-

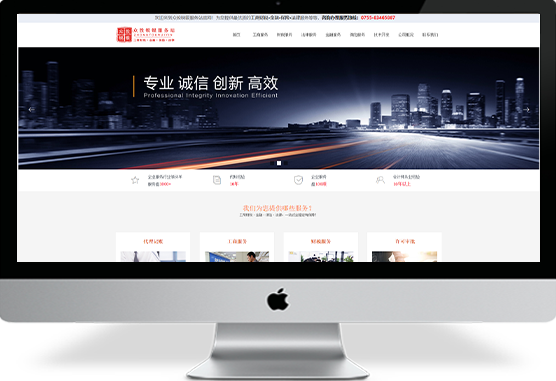
啊啊啊啊啊片
聚为科技为您呈现新的企业营销型网站建设成功案例、集团网站建设案例、品牌网站建设案例、外贸营销型网站案例等,湖北聚为科技已帮助500+家企业实现网上盈利,他们在互联网领域已成为行业翘楚,咨询热线:400-806-4100¥ 0.00立即购买
-
污污污男生爆操嫩逼免费看网站
湖北聚为科技是一家懂营销的互联网+网站设计定制公司,打造富有视觉冲击力、营销说服力和利于SEO优化排名的企业官方网站建设。更多网站建设案例和网站改版案例分享欢迎访问湖北聚为科技官网。¥ 0.00立即购买
-

骚逼操av
早在几年前,使用大图片或者全屏背景的网页设计已经成为一种趋势,直到如今,这样的网页设计风格依旧备受欢迎,因为这样的设让网页外观看起来非常的简约大方。¥ 0.00立即购买
-
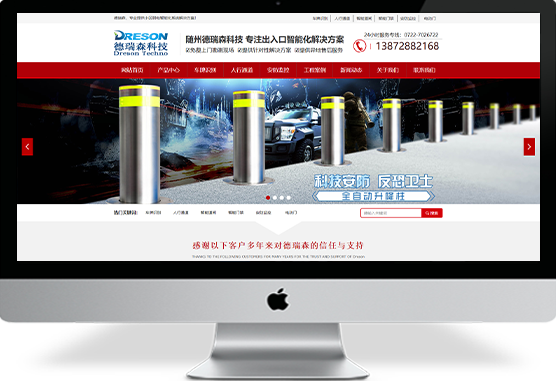
大黑屌视频fuck
聚为科技5多年专注于网站建设,模板网站、定制网站、H5网站、个性化开发平台等,拥有丰富的企业网站建站经验,近年来制作并完成了许多专业精美的网站建设案例,网站制作案例,网站设计案例等¥ 0.00立即购买
-

欧美操大逼片
优秀网站制作公司聚为科技目前有1000多家优秀的网站设计案例,有2000多家品牌网站建设案例,是一家技术实力超强的网站设计公司,在同行里有很高的知名度¥ 0.00立即购买
-
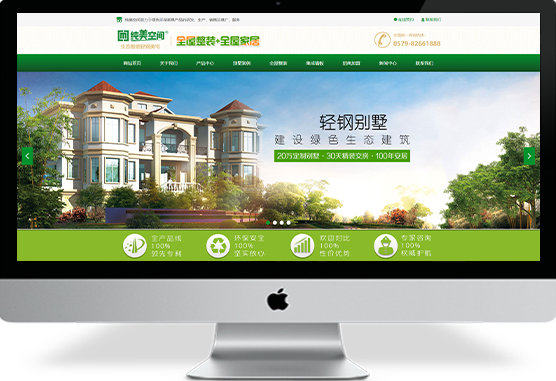
www涩
企业官网制作:特价促销:原价3980元,现价3280元!解决方案基础应用产品服务1.赠送国际域名一个(.com/.net)或国内域名(.cn)2.在我司购买域名以及独立IP空间,200M空间可获赠100M企业邮箱网站建设3.网站美工设计(专业美工DIV+CSS设计)a、改善网页内容布局,改善网站架构¥ 0.00立即购买
最新的资讯动态以及建站相关知识
网站建设知识/小程序开发知识/网页设计观点/优化推广知识,将想法与焦点与您一起分享
-
-
随县福润养殖项目网上二次公示美女操鸡巴粉嫩嫩av
넶66 2024-04-19
- 2024-04-19
- 2024-04-19
- 2024-04-19
- 2024-04-19
- 2024-04-19
-
-
- 2024-04-19
- 2024-04-19
- 2024-04-19
- 2024-04-19
- 2024-04-19
- 2024-04-19
- 2024-04-19
- 2024-04-19
- 2024-04-19
- 2024-04-19
-
- 2024-04-19
- 2024-04-19
- 2024-04-19
- 2024-04-19
- 2024-04-19
- 2024-04-19
- 2024-04-19
- 2024-04-19
- 2024-04-19
- 2024-04-19
-
-
2024-04-192024-04-192024-04-192024-04-192024-04-192024-04-192024-04-192024-04-192024-04-192024-04-192024-04-192024-04-192024-04-192024-04-192024-04-192024-04-192024-04-19ꂀ 虚拟主机知识大全2024-04-192024-04-192024-04-19
ꁲ
您好,聚为科技有什么可以帮您的吗?
网站建设?微信小程序开发?电商网站建设?app开发?
您也可以直接电话沟通:
0722-7096888
售后服务:17607167278(同微信)
联系地址:湖北省随州市曾都区人才创新创业孵化基地6楼6088室
男人桶女人的逼-欧美熟妇黑BBXX老妇 ✅ 免费看片SE356.com Copyright © 2015-2020 湖北聚为科技股份有限公司 版权所有 版权声明:部分图片及内容来源于网络,版权归原作者所有,如有侵权请告知我们删除
友情链接(欢迎网站建设公司及相关行业网站与聚为科技交换链接!链接交换QQ:295616443)
随州网站建设、网站设计制作公司-聚为科技,已为众多企业提供网站建设 网站制作 响应式网站 手机网站建设 虚拟主机 云主机 服务器租用 网站托管等建站解决方案
-
ꁸ 回到顶部
-
ꂅ 0722-7096888
-
ꁗ QQ咨询
-
ꀥ 微信咨询